Sometimes one needs to simulate a Poisson point process on a disk. I know I often do. A disk or disc, depending on your spelling preference, is isotropic or rotationally-invariant, so a lot of my simulations of a Poisson point process happen in a circular simulation window when I am considering such a setting. For example, maybe you want to consider a single wireless receiver in a Poisson network of wireless transmitters, which only cares about the distance to a transmitter. Alternatively, maybe you want to randomly sprinkle a virtual cake. What to do? A Poisson point process on a disk is the answer.
In this post I will simulate a Poisson point process with intensity \(\lambda>0\) on a disk with radius \(r>0\).
Steps
The simulation steps are very similar to those in the previous post where I simulated a homogeneous Poisson point process on a rectangle, and I suggest going back to that post if you are not familiar with the material. The main difference between simulation on a rectangle and a disk is the positioning of the points, but first we’ll look at the number of points.
Number of points
The number of points of a Poisson point process falling within a circle of radius \(r>0\) is a Poisson random variable with mean \(\lambda A\), where \(A=\pi r^2\) is the area of the disk. As in the rectangular case, this is the most complicated part of the simulation procedure. But as long as your preferred programming language can produce (pseudo-)random numbers according to a Poisson distribution, you can simulate a homogeneous Poisson point process on a disk.
To do this in MATLAB, use the poissrnd function with the argument \(\lambda A\). In R, it is done similarly with the standard function rpois . In Python, we can use either the scipy.stats.poisson or numpy.random.poisson function from the SciPy or NumPy libraries.
Locations of points
We need to position all the points randomly and uniformly on a disk. In the case of the rectangle, we worked in Cartesian coordinates. It is then natural that we now work in polar coordinates. I’ll denote the angular and radial coordinate respectively by \(\theta\) and \(\rho\). To generate the random angular (or \(\theta\)) values, we simply produce uniform random variables between zero and one, which is what all standard (pseudo-)random number generators do in programming languages. But we then multiply all these numbers by \(2\pi\), meaning that all the numbers now fall between \(0\) and \(2\pi\).
To generate the random radial (or \(\rho\)) values, a reasonable guess would be to do the same as before and generate uniform random variables between zero and one, and then multiply them by the disk radius \(r\). But that would be wrong.
Before multiplying uniform random variables by the radius, we must take the square root of all the random numbers. We then multiply them by the radius, generating random variables between \(0\) and \(r\). (We must take the square root because the area element of a sector or disk is proportional to the radius squared, and not the radius.) These random numbers do not have a uniform distribution, due to the square root, but in fact their distribution is an example of the triangular distribution, which is defined with three real-valued parameters \(a\), \(b\) and \(c\), and for our case, set \(a=0\) and \(b=c=r\).
In summary, if \(U\) and \(V\) are two independent uniform random variables on \((0,1)\), then random point located uniformly on a disk of radius \(r\) has the polar coordinates \((r\sqrt(U), 2\pi V)\).
From polar to Cartesian coordinates
That’s it. We have generated polar coordinates for points randomly and uniformly located in a disk. But to plot the points, often we have to convert coordinates back to Cartesian form. This is easily done in MATLAB with the pol2cart function. In languages without such a function, trigonometry comes to the rescue: \(x=\rho\cos(\theta)\) and \(y=\rho\sin(\theta)\).
Equal x and y axes
Sometimes the plotted points more resemble points on an ellipse than a disk due to the different scaling of the x and y axes. To fix this in MATLAB, run the command: axis square. In Python, set axis(‘equal’) in your plot; see this page for a demonstration.
Code
I’ll now give some code in MATLAB, R and Python, which, as you can see, are all very similar
MATLAB
%Simulation window parameters
r=1; %radius of disk
xx0=0; yy0=0; %centre of disk
areaTotal=pi*r^2; %area of disk
%Point process parameters
lambda=100; %intensity (ie mean density) of the Poisson process
%Simulate Poisson point process
numbPoints=poissrnd(areaTotal*lambda);%Poisson number of points
theta=2*pi*(rand(numbPoints,1)); %angular coordinates
rho=r*sqrt(rand(numbPoints,1)); %radial coordinates
%Convert from polar to Cartesian coordinates
[xx,yy]=pol2cart(theta,rho); %x/y coordinates of Poisson points
%Shift centre of disk to (xx0,yy0)
xx=xx+xx0;
yy=yy+yy0;
%Plotting
scatter(xx,yy);
xlabel('x');ylabel('y');
axis square;
R
Note: it is a bit tricky to write “<-” in the R code (as it automatically changes to the html equivalent in the HTML editor I am using), so I have usually used “=” instead of the usual “<-”.
#Simulation window parameters r=1; #radius of disk xx0=0; yy0=0; #centre of disk areaTotal=pi*r^2; #area of disk #Point process parameters lambda=100; #intensity (ie mean density) of the Poisson process #Simulate Poisson point process numbPoints=rpois(1,areaTotal*lambda);#Poisson number of points theta=2*pi*runif(numbPoints);#angular of Poisson points rho=r*sqrt(runif(numbPoints));#radial coordinates of Poisson points #Convert from polar to Cartesian coordinates xx=rho*cos(theta); yy=rho*sin(theta); #Shift centre of disk to (xx0,yy0) xx=xx+xx0; yy=yy+yy0; #Plotting par(pty="s") plot(xx,yy,'p',xlab='x',ylab='y',col='blue');
Of course, with the amazing spatial statistics library spatstat, simulating a Poisson point process in R is even easier.
library("spatstat"); #load spatial statistics library
#create Poisson "point pattern" object
ppPoisson=rpoispp(lambda,win=disc(radius=r,centre=c(xx0,yy0)))
plot(ppPoisson); #Plot point pattern object
#retrieve x/y values from point pattern object
xx=ppPoisson$x;
yy=ppPoisson$y;
Actually, you can even do it all in two lines: one for loading the spatstat library and one for creating and plotting the point pattern object.
Python
Note: “lambda” is a reserved word in Python (and other languages), so I have used “lambda0” instead.
import numpy as np; #NumPy package for arrays, random number generation, etc
import matplotlib.pyplot as plt #for plotting
#Simulation window parameters
r=1; #radius of disk
xx0=0; yy0=0; #centre of disk
areaTotal=np.pi*r**2; #area of disk
#Point process parameters
lambda0=100; #intensity (ie mean density) of the Poisson process
#Simulate Poisson point process
numbPoints = np.random.poisson(lambda0*areaTotal);#Poisson number of points
theta=2*np.pi*np.random.uniform(0,1,numbPoints); #angular coordinates
rho=r*np.sqrt(np.random.uniform(0,1,numbPoints)); #radial coordinates
#Convert from polar to Cartesian coordinates
xx = rho * np.cos(theta);
yy = rho * np.sin(theta);
#Shift centre of disk to (xx0,yy0)
xx=xx+xx0; yy=yy+yy0;
#Plotting
plt.scatter(xx,yy, edgecolor='b', facecolor='none', alpha=0.5 );
plt.xlabel("x"); plt.ylabel("y");
plt.axis('equal');
Julia
After writing this post, I later wrote the code in Julia. The code is here and my thoughts about Julia are here.
Results
MATLAB 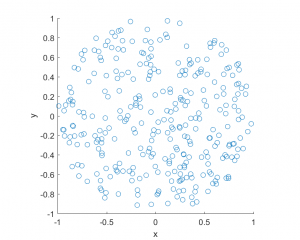
R 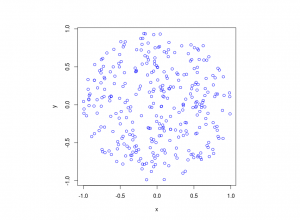
Python
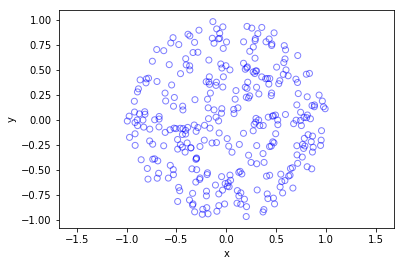
Further reading
The third edition of the classic book Stochastic Geometry and its Applications by Chiu, Stoyan, Kendall and Mecke details on page 54 how to uniformly place points on a disk, which they call the radial way. The same simulation section appears in the previous edition by Stoyan, Kendall and Mecke (Chiu didn’t appear as an author until the current edition), though these books in general have little material on simulation methods. There is the book Spatial Point Patterns: Methodology and Applications with R written by spatial statistics experts Baddeley, Rubak and Turner, which covers the spatial statistics (and point process simulation) R-package spatstat.



Can you please tell how to generate voronoi diagram in Matlab with centers generated according to poisson point process and then users are substituted in each voronoi cell again according to poisson point process. If matlab code could be provided then it would be of great help. Thanks in advance.
MATLAB already has an inbuilt function for this:
https://au.mathworks.com/help/matlab/ref/voronoi.html
I recommend you look at their examples first. I have code that implements MATLAB’s voronoi function and does something similar to you what you want, but it needs tidying up. Perhaps I’ll post it sometime in the near future.
For the second part, I believe a Poisson point process over the entire simulation window/region corresponds to an (individual) independent Poisson point process in each Voronoi window. This should be true because of a defining property of the Poisson point process, which is sometimes called the scattering property (ie the number of points of a Poisson point process in non-overlapping/disjoint regions are independent variables.) This will NOT be true if it’s not a Poisson point process.
In short, just simulate another Poisson point process over the entire region for the users (it can have a intensity different from the first Poisson point processed used to create the Voronoi tessellation/diagram).
Hi Paul,
Many thanks for your posts.
Please consider the event “only the nearest point to the center of the disc, is displaced in a random direction, independently from the other points”. Can you please tell if the new point process remains a PPP.
Hi Yassine,
Doubtful. You are changing the homogeneous PPP in the region around the center of the disk, where there is now *less* or *more* chance (depending on the random displacement) of finding a point. Although perhaps the change is very small.
The only way I see it still being a PPP is by positioning it with the same random distribution that it was originally positioned with: (in polar coordinates) the angular variable $\Theta$ is uniform on $(0,2\pi)$ and the radial component $R$ has a Rayleigh distribution $P(R\leq r) = 1- exp(- \lambda\pi r^2 )$ (which is the distribution of the nearest-neighbour distance ). But then you haven’t changed the Poisson point process (in distribution) as it statistically behaves the same.
This code supports my claim:
numbSim=10^4; %number of simulations
%Simulation window parameters
r=1; %radius of disk
xx0=0; yy0=0; %centre of disk
areaTotal=pi*r^2; %area of disk
%Point process parameters
lambda=5; %intensity (ie mean density) of the Poisson process
countPointsMid=zeros(numbSim,1);
rMid=r/2; %arbitrary point along the radius
%loop through for many simulations
for ss=1:numbSim
%Simulate Poisson point process
numbPoints=poissrnd(areaTotal*lambda);%Poisson number of points
theta=2*pi*(rand(numbPoints,1)); %angular coordinates
rho=r*sqrt(rand(numbPoints,1)); %radial coordinates
[rhoMin, indexMin]=min(rho);
%change rho and theta -- comment next 4 commands for regular Poisson
%generate
thetaNew=2*pi*rand(1);
rhoNew=r*rand(1); %uniform rho
%rhoNew=sqrt(-log(rand(1))/lambda/pi); %Rayleigh rho
%replace new variables
theta(indexMin)=thetaNew;
rho(indexMin)=rhoNew;
countPointsMid(ss)=sum(rho
how can we simulate a poisson point rocess in R^3
Hi Hawat,
You would need to do it in a cube or rectangular prism. It’s just a variation of doing it on a rectangle, but you use the volume (instead of the area) of the cuboid (or rectangular prism). And then for each point you generate a third uniform random variable Z for the three Cartesian coordinate. Look at this post, where the Poisson point process is simulated on a rectangle:
https://hpaulkeeler.com/poisson-point-process-simulation/
I have included a section at the end detailing how to do it in a bounded three-dimensional box (that is, cuboid or rectangular prism).
Alternatively, you could also do it in a 3-D sphere. This post does it for a sphere (the *surface* of a ball), but I detail at the end of the post how you can do position points uniformly *inside* a ball:
https://hpaulkeeler.com/simulating-a-poisson-point-process-on-a-sphere/
Hi Paul!
I have a two-tier network comprising devices, gateways and the base station. Gateways are distributed according to PPP while the devices themselves are distributed according MCP. How do I achieve this in MATLAB assuming a circular area of coverage for each gateway generated, within which the respective devices are distributed wrt MCP?
Hi Mots’oane,
By MCP, I assume you mean Matern cluster process. I would suggest reading this post:
https://hpaulkeeler.com/simulating-a-matern-cluster-point-process/
Then you just have to modify code that simulates the underlying Poisson point process, so the points are scattered on a disk, like they are in this post.
Sir, thank you for sharing this article. I have used the MATLAB code to generate a base station PPP inside B(0,R) where each BS has a mobile user at a fixed distance ‘r’ from its associated BS. To define mobile users, I have used
rhoM = rho + r;
thetaM = theta + unifrnd(0, 2*pi);
After that I have used pol2cart operator to get the coordinates of the mobile user but I am not getting the required result. May you suggest anything regarding this problem?
I am also thinking to generate a high density mobile user point process separatly and then used thing process to get the target mobile user the given fixed distance. Can I simulate this problem in this way?
Thanks and Regards
I don’t understand what you’re doing. In polar coordinates, you cannot simply add the radial (rho) and angular (theta) components. You do that in Cartesian coordinates. If you look at my MATLAB code, that’s what I do:
https://github.com/hpaulkeeler/posts/blob/master/PoissonDisk/PoissonDisk.m
Sir, thank you for sharing this article. I have used the MATLAB code to generate a base station PPP inside B(0,R) where each BS has a mobile user at a fixed distance ‘r’ from its associated BS. To define mobile users, I have used
rhoM = rho + r;
thetaM = theta + unifrnd(0, 2*pi);
After that I have used pol2cart operator to get the coordinates of the mobile user but I am not getting the required result. May you suggest anything regarding this problem?
I am also thinking to generate a high density mobile user point process separatly and then used thining to get the target mobile user the at the fixed distance ‘r’. Can I simulate this problem in this way?
Thanks and Regards
Hello Prof. many thanks for sharing these codes
I assumed I have BS at the center of the the circle and users should be deployed at least 35m from the BS. as well as I assumed this circle is compromised of two circles, which means there are inner cercle used for deplying BS-users and out circle used to deply another users. So, can I impement something like that assumtion ?
Hi, sir. Thanks for your sharing this article. It’s helpful to simulate PPP in a disk.
I also have a question about the simulation. In PPP, the number of points is a variable, maybe we should not use the mean of the variable to replace the points number. In each cycle of executing the codes, the number of points should be different.
Thank you for your comment. In the code, the number of points is a random variable. It changes every simulation. It is not a constant.
But in the code you pass a constant (the mean) to a function that generates Poisson random variables (or more correctly, variates). In other words, the input is constant, but the output is random. To do this in MATLAB, use the poissrnd function with the argument λA. In R, it is done similarly with the standard function rpois. In Python, you can use either the scipy.stats.poisson or numpy.random.poisson function from the SciPy or NumPy libraries.
Thanks for your detailed interpretation with patience, Sir. It’s really helpful to my work.
No problem.
Hi ,Sir, thanks for publishing this helpful article, but I have a problem, I made a simulation for uplink system in square and I want to make the same on disk but the results are not the same, is the shape changes the result.
The results will be different due to “edge effects” (you are approximating the infinite plane with a square or disk). But the results should be the same as the disk and square grow in size.
Hi Paul,
Thanks for your helpful articles!
I have a two tiers wireless network , drones and ground base stations, both are distributed according to BPP in a circular area with radius R. With a number of users distributed randomly on the ground. How to simulate in MATLAB please? Your help is really appreciated.
Thank you for your comment. For the drones (or base stations), you just need to simulate the binomial point process on a disk. You take the the simulation method for a Poisson point process on a disk and replace the random (Poisson) variable of points, say $N$, with a constant number of points, say $n$.
See the posts:
https://hpaulkeeler.com/simulating-a-poisson-point-process-on-a-disk/
https://hpaulkeeler.com/binomial-point-process/
Here is the corresponding code for the two above posts:
https://github.com/hpaulkeeler/posts/tree/master/PoissonDisk
https://github.com/hpaulkeeler/posts/tree/master/BinomialRectangle